Blog
You may or may not find something useful here.
Browse the archive to see all entries.
I recently bought a new Radeon RX 7800 XT, mainly to use the OpenCL features, for example with Darktable.
However, I found it not easy to install all the drivers - especially the OpenCL created some headache for me. Therefore is here a short how to. I used Debian Bookworm for this.
- First of all, install the firmware-amd-graphics package. Note:
There is still this bug-report Debian bug #1052714, which means you have to
download fresh firmware from the kernel firmware repo.
To do so, download the latest tar (I used linux-firmware-20230919.tar.gz, copy the folder
amdgputo/usr/lib/firmwareand runupdate-initramfs -c -k all) - Now - as described in the Debian Wiki Page, remove all nvidia stuff you might have:
apt purge '*nvidia*' - Install the required packages:
apt install libgl1-mesa-dri libglx-mesa0 mesa-vulkan-drivers xserver-xorg-video-all - Reboot. At least the graphical user interface should work now. You should also be able to play games. But OpenCL will not work.
- For OpenCL, I found that the mesa driver (mesa-opencl-icd) does not work. You need the AMD "ROCm" stuff.
- To install it, add the following repository:
deb [arch=amd64] https://repo.radeon.com/rocm/apt/5.7 focal main, then runapt updateand installapt install rocm-opencl-runtime - Add your user to the render and video groups:
sudo usermod -a -G render,video $LOGNAME(This is very important! If you do not do this, OpenCL will print weird error messages but not say that you are not member of the group...) - Reboot and you should see OpenCL working, when using
darktable-cltest
Here are some darktable benchmarks with my hardware (Ryzen 9 3900, 64GB RAM, SATA SSD, RX 7800 XT):
[dt_opencl_device_init]
DEVICE: 0: 'gfx1101'
PLATFORM NAME & VENDOR: AMD Accelerated Parallel Processing, Advanced Micro Devices, Inc.
CANONICAL NAME: amdacceleratedparallelprocessinggfx1101
DRIVER VERSION: 3590.0 (HSA1.1,LC)
DEVICE VERSION: OpenCL 2.0
DEVICE_TYPE: GPU
GLOBAL MEM SIZE: 16368 MB
MAX MEM ALLOC: 13913 MB
MAX IMAGE SIZE: 16384 x 16384
MAX WORK GROUP SIZE: 256
MAX WORK ITEM DIMENSIONS: 3
MAX WORK ITEM SIZES: [ 1024 1024 1024 ]
ASYNC PIXELPIPE: NO
PINNED MEMORY TRANSFER: NO
MEMORY TUNING: NO
FORCED HEADROOM: 400
AVOID ATOMICS: NO
MICRO NAP: 250
ROUNDUP WIDTH: 16
ROUNDUP HEIGHT: 16
CHECK EVENT HANDLES: 128
PERFORMANCE: 23.375
TILING ADVANTAGE: 0.000
DEFAULT DEVICE: NO
KERNEL BUILD DIRECTORY: /usr/share/darktable/kernels
KERNEL DIRECTORY: /home/reox/.cache/darktable/cached_v1_kernels_for_AMDAcceleratedParallelProcessinggfx1101_35900HSA11LC
CL COMPILER OPTION: -cl-fast-relaxed-math
KERNEL LOADING TIME: 0.0382 sec
===================
# OpenCL TEST #
===================
took 0.968 secs
took 0.966 secs
took 0.961 secs
===================
# CPU ONLY TEST #
===================
took 8.705 secs
took 8.722 secs
took 8.775 secs
If you use piwigo you might have noticed that EXIF is not working properly sometimes. Especially if you have multiple cameras and want to extract the lens data.
The reason for this is manyfold:
- Each Camera might use a different EXIF tag for the lens
- The lens might not even be written correctly in the EXIF data
- the PHP implementation that piwigo uses cannot correclty extract the required tag
At least for the first issue, I found a solution. Simply extract all possible tags and then merge them with this personal plugin:
<?php
/*
Plugin Name: EXIF Merge
Version: 1.0
Description: it merges EXIF tags
Plugin URI: -
Author: Sebastian Bachmann
Author URI: https://reox.at
*/
add_event_handler('format_exif_data', 'exif_merge');
/**
* EXIF Merge.
*
* Merges some keys in the EXIF Data, so we have always a correct lens description
*
* @param $exif dictionary with EXIF keys and values
* @return merged array
*/
function exif_merge($exif) {
if (is_array($exif)) {
// 5DmkII stores lens in UndefinedTag:0x0095
if (array_key_exists('UndefinedTag:0x0095', $exif)) {
$exif['UndefinedTag:0xA434'] = $exif['UndefinedTag:0x0095'];
unset($exif['UndefinedTag:0x0095']);
}
}
return $exif;
}
?>
In the settings, enable EXIF and add the following fields:
$conf['show_exif'] = true;
$conf['show_exif_fields'] = array(
'FocalLength',
'ExposureTime',
'ISOSpeedRatings',
'FNumber',
'Model', // Camera Model
'UndefinedTag:0xA434', // lens
'UndefinedTag:0x0095', // lens tag for 5Dmkii
);
In addition, you might also want to use exif_view.
To check which tags are used and known by PHP, you can use the tools/metadata.php script. You have to change the filename to inspect inside the script though.
Sometimes you need to display tensors in paraview, and for that reason the VTK ASCII file format is perfect. You can easily write this by hand even! However, the documentation is bad on it and it took me quite some time to figure out how to write tensors by hand, thus here is an explanation for the future - when I again need it and cannot find the documentation...
The VTK format knows two kinds of notations: full 2nd rank tensors and symmetric 2nd rank tensors. A 2nd rank tensor can be written as a matrix with its components like this:
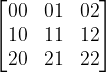
Now, the ordering in the VTK file for TENSOR (full tensor) is:
00 01 02 10 11 12 20 21 22
The ordering for the TENSOR6 (symmetric tensor) is:
00 11 22 01 12 02
To see this in action, here are two equivalent files:
# vtk DataFile Version 2.0
HMA Analysis
ASCII
DATASET UNSTRUCTURED_GRID
POINTS 1 float
1.23 2.3556 3.4525
POINT_DATA 1
TENSORS6 OrientTensor float
1.5 1.8 2.0 2.5 0.8 3.4
# vtk DataFile Version 2.0
HMA Analysis
ASCII
DATASET UNSTRUCTURED_GRID
POINTS 1 float
1.23 2.3556 3.4525
POINT_DATA 1
TENSORS OrientTensor float
1.5 2.5 3.4
2.5 1.8 0.8
3.4 0.8 2.0
Note: You can also add tensors to CELL_DATA, but then they can only be
displayed in paraview if you use the Cell Data to Point Data filter.
Many modern cameras allow for automated focus stacking (focus bracketing in Canon speech).
A quick excursion how this works on a Canon RP (and similar R models):
- Enable Focus-Bracketing
- Set the maximum number of images, i.e. 40 or more for macros - for landscape it will often only be 10 images that are taken. The camera will decide anyway when to stop (When focus reaches infinity), thus you can leave this on a high value just to be sure! Macro images require more photos than landscape (my rule of thumb...)
- You can adjust the focus increment, for me 4 is fine. You might decrease it for smaller aperture numbers.
- Now, focus on the nearest distance you want to have sharp in the image
- Maybe enable 2 second timer to reduce vibrations on the tripod and press the shutter button once.
- On a bright day, this takes only a second to gather all the images!
Using enfuse and the tools from hugin-tools, it is quite easy to create a nice focus stacked image out of the set of images. These commands came from foto.schwedenstuhl.de.
I use darktable to collect my photos, so the first step is to export the set of images as .tiff files.
In the next step, we use the tool align_image_stack from hugin-tools to align the images:
align_image_stack -v -m -a Aligned *.tif
It seems counter-intuitive to do the alignment, even if we used a tripod. But there is a reason for this: during focusing, the actual focal length shifts a tiny bit (This phenomena is also called "focus breathing"). For landscape photography, this might not necessarily be required (I tested it). But still, you can see a small effect of this even in landscape images. For macro-photos, this matters a lot!
Now, we can call enfuse to create the fused image:
enfuse \
--exposure-weight=0 \
--saturation-weight=0 \
--contrast-weight=1 \
--hard-mask \
--contrast-window-size=9 \
--output=fused.tif \
Aligned*.tif
You can play around with the options, but these were already recommended elsewhere and seem to be a good starting point.
Here is an example. This is the first image of the stack, notice the very shallow focus:
Now, the focus stacked image - you can clearly see that the focus is over the full depth!
I kept a lot of lab notes in Word documents. For each day I wrote notes, I used a header with the date and put the text below. Recently, I switched to do notes in LogSeq, and now I wanted to convert all the existing notes into markdown files.
There are a few things that make this complicated:
- pandoc can do this, but only writes a single markdown file
- pandoc further writes images all in the same folder with a name
like
image1.png, which makes it very hard to run it on multiple files
So, here are some scripts to circumvent this.
First, we use a LUA filter to rename the image files attached to the docx by their SHA1 (a function that is luckily supplied with pandoc!):
-- Basic concept from https://stackoverflow.com/a/65389719/446140
-- CC BY-SA 4.0 by tarleb
--
-- Adjusted by reox, released as CC BY-SA 4.0
local mediabag = require 'pandoc.mediabag'
-- Stores the replaced file names for lookup
local replacements = {}
-- Get the extension of a file
function extension(path)
return path:match("^.+(%..+)$")
end
-- Get the directory name of a file
function dirname(path)
return path:match("(.*[/\\])")
end
-- Delete image files and re-insert under new name
-- Uses the SHA1 of the content of the file to rename the image path
for fp, mt, contents in mediabag.items() do
mediabag.delete(fp)
local sha1 = pandoc.utils.sha1(contents)
-- Stores the file in the same name as specifed
--local new_fp = dirname(fp) .. "/image_" .. sha1 .. extension(fp)
-- Stores them in a folder called assets
local new_fp = "assets" .. "/image_" .. sha1 .. extension(fp)
replacements[fp] = new_fp
mediabag.insert(new_fp, mt, contents)
end
-- adjust path to image file
function Image (img)
-- Use this to simply replace
-- img.src = replacements[img.src]
-- Move into relative folder
img.src = "../" .. replacements[img.src]
-- Set a title, similar to logseq
img.caption = "image.png"
-- Remove attributes such as width and height
img.attr = {}
return img
end
Some specialities are also that all attributes are removed from the image, the
caption is set to image.png and the files are also moved into a folder named
assets - like used in logseq.
Now, we can convert the docx to markdown using pandoc:
pandoc -f docx -t markdown notebook.docx -o full_file.md --extract-media=. --lua-filter=filter.lua
For the splitting part, we can use the tool csplit, contained in coreutils.
Basically, we tell it to split the file at every occurence of a H3:
csplit --prefix='journals/journal_' --suffix-format='%03d.md' full_file.md '%^### %' '{1}' '/^### /' '{*}'
An additional adjustment is to skip everything up to the first H3.
Everything can be put into a small script to convert docx files, put them into the right folder and also rename them into journal files, logseq understands:
#!/bin/bash
convert_file () {
pandoc -f docx -t markdown "$1" -o full_file.md --extract-media=. --lua-filter=mediabag_filter.lua
#
# Remove H1 and H2 from file
sed full_file.md -e 's/^[#]\{1,2\} .*$//' > filtered.md
# Remove empty headers. This seems to happen when we have two lines of H3 in the
# file by acciedent
sed -i filtered.md -e 's/^#\+\s\+$//'
# Split file
mkdir -p journals
# Skip any text up to the first H3, then split at each H3
csplit --prefix='journals/journal_' --suffix-format='%03d.md' filtered.md '%^### %' '{1}' '/^### /' '{*}'
# Rename files
for file in journals/journal_*.md; do
h=$(head -n 1 "$file")
if ! [ "x${h:0:4}" = "x### " ]; then
echo "$file does not start with header!"
exit 1
fi
Y=${h:10:4}
M=${h:7:2}
D=${h:4:2}
dir=$(dirname "$file")
echo "$file --> $dir/${Y}_${M}_${D}.md"
# Remove header line, not required by logseq
sed -i "$file" -e "1d"
# Shift headers by 3
# Apply table format, that logseq understands
pandoc -f markdown -t markdown-simple_tables-grid_tables-multiline_tables+pipe_tables "$file" --shift-heading-level-by=-3 -o "$dir/${Y}_${M}_${D}.md"
rm $file
done
# Cleanup
rm full_file.md
rm filtered.md
}
for arg in "$@"; do
echo "Converting $arg ..."
convert_file "$arg"
done
The only thing that does not work properly is to automatically convert the files into the list format that logseq uses... That is tricky to do, because we cannot simply use every paragraph as a block.
Furthermore, some equations do not work properly and some other text looks terrible. However, this is only a start and you can probably improve this script quite a bit and also adopt it to your needs.
Yet another fun exercise to calculate something!
Pressure sensors measure the pressure on their altitude, but we eventually want the pressure at sea-level, for example to check with the weather forecast.
That can be done, by using some equations, if we have a sensor that can measure temperature, humidity and pressure (for example a BME280):
template:
- sensor:
- name: "Pressure sea level"
unique_id: "pressure_red"
state: |-
{%- set g0 = 9.80665 %}
{%- set RG = 287.05 %}
{%- set Ch = 0.12 %}
{%- set a = 0.0065 %}
{%- set h = 162.0 %}
{%- set p = states('sensor.outdoor_pressure') | float %}
{%- set phi = 0.01 * (states('sensor.outdoor_humidity') | float) %}
{%- set t = (states('sensor.outdoor_temperature') | float) %}
{%- set T = 273.15 + t %}
{%- set E = phi * 6.112 * e**((17.62 * t) / (243.12 + t)) %}
{%- set x = (g0 / (RG * (T + Ch * E + a * (h / 2)))) * h %}
{{ (p * e**x) | round(1) }}
icon: "mdi:gauge"
unit_of_measurement: 'hPa'
state_class: "measurement"
device_class: "pressure"
The only thing you have to set is the height (h) in meters.
If you have a temperature and humidity sensor, you can calculate the dewpoint using the Magnus formula. See for example the german Wikipedia.
It is pretty straight forward to create a template sensor out of this:
template
- sensor:
- name: "Dew Point"
unique_id: "indoor_dewpoint"
state: |-
{%- set t = states('sensor.indoor_temperature') | float %}
{%- set lnrh = log(0.01 * states('sensor.indoor_humidity') | float) %}
{%- set k2 = 17.62 %}
{%- set k3 = 243.12 %}
{%- set q1 = (k2 * t) / (k3 + t) %}
{%- set q2 = (k2 * k3) / (k3 + t) %}
{{ (k3 * ((q1 + lnrh) / (q2 - lnrh))) | round(1) }}
unit_of_measurement: "°C"
icon: "mdi:thermometer-water"
device_class: "temperature"
state_class: "measurement"
I use k9-mail on Android. It works perfectly fine, except for one thing: it does not support yearly based archives... However, k9-mail supports archives in general, thus you can easily set an archive folder and move the messages there.
That's were IMAPSIEVE comes into play - at least I thought so on first glance. Then I discovered, that it is actually not that easy to configure, as the documentation is very sparse. But fortunately, I got it working and want to document the steps I took.
For user and mailbox specific scripts, you also need to enable IMAP METADATA. Thus, the first step is to enable that.
Then you can enable IMAPSIEVE. The required parts are (for me on Debian):
in /etc/dovecot/conf.d/20-imap.conf set:
protocol imap {
# ...
mail_plugins = $mail_plugins imap_sieve
# ...
}
and in /etc/dovecot/conf.d/90-sieve.conf set:
plugin {
# This option should already be set when you configured sieve...
sieve = file:~/sieve;active=~/.dovecot.sieve
# ...
# These two are the important bits for imapsieve:
sieve_plugins = sieve_imapsieve
imapsieve_url = sieve://your_managesieve_server:4190
# ...
}
I simply assume that you have already configured managesieve and sieve correctly
and to your needs. The imapsieve_url seems to be important and you have to
configure the server URL to managesieve here. AFAIK it is only there to hint you
the managesieve url though...
Now that you have configured all the important bits, you can test if the things
are enabled. Run openssl s_client to connect to your IMAP like:
$ openssl s_client -crlf -connect your_mail_server:993
you can login with
a login your_username your_password
now, you should get a list of the CAPABILITIES of the server. Check that in the
list there is METADATA and IMAPSIEVE=sieve://your_managesieve_server:4190.
Now, you can set mailbox specific sieve scripts, using the IMAP commandline.
The important command here is SETMETADATA and GETMETADATA to verify.
Information about these commands can be found in the RFC 5464.
Basically, you need to set the key /shared/imapsieve/script with the sieve
script as value, per mailbox you want to filter.
For example, if you have uploaded a sieve script called archivefilter (this will
physically be stored in %h/sieve/archivefilter.sieve if you configured the
sieve option as I did), then you need to type the following command to
activate it for the mailbox Archives:
a SETMETADATA "Archives" (/shared/imapsieve/script "archivefilter")
You can verify that by typing:
a GETMETADATA "Archives" /shared/imapsieve/script
if you are done, logout:
a LOGOUT
Now for the sieve script itself. I want to redirect all messages which are moved
into the Archives folder to Archives.YYYY - so that I can use the archives
setting from k9-mail but keep my yearly archive.
So far, I was able to solve this using this sieve script:
require ["imapsieve", "environment", "date", "fileinto", "mailbox", "variables"];
# Rule for imapsieve, move messages from Archives to Archives.YEAR
if anyof (environment :is "imap.cause" "APPEND", environment :is "imap.cause" "COPY") {
if date :matches "received" "year" "*" {
fileinto :create "Archives.${1}";
stop;
}
# No received date available... odd but OK... sort by currentdate:
# (Not sure though if this can actually happen?)
if currentdate :matches "year" "*" {
fileinto :create "Archives.${1}";
stop;
}
}
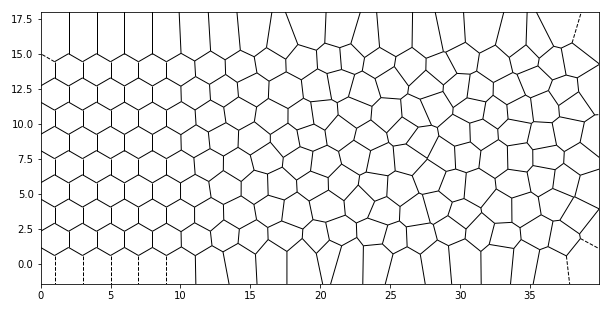
Somewhere I saw a picture of a high-performance computer, where the doors would have a hexagonal pattern that would "degrade" into a irregular Voronoi pattern as ventilation holes.
That can be replicated quite easily with a short python script! If you like, you can then use my Voronoi pattern generator for FreeCAD to build such ventilation holes yourself.
The python code is pretty straight forward. First, you need a couple of libraries:
import numpy as np
from scipy.spatial import Voronoi, voronoi_plot_2d
import matplotlib.pyplot as plt
from itertools import count
Next up, we generate a couple of points that are the centers of tiled hexagons:
def hex_pattern(a: float = 1, nx: int = 20, ny: int = 10, start: int = 1) -> np.array:
"""
Create a pattern with nx times ny cells
:param float d: edge length of the hexagon
:param int nx: number of cells in x-direction
:param int ny: number of cells in y-direction
:param int start: if the offset is on even (0) or odd (1) rows
"""
points = np.empty((nx * ny, 2), dtype=float)
if start not in (0, 1):
raise ValueError('start must be either 0 or 1')
if nx < 1:
raise ValueError("there must be a minimum of 1 cells in x")
if ny < 1:
raise ValueError("there must be a minimum of 1 cells in y")
if a <= 0:
raise ValueError("Edge length of hexagon must be positive and non-zero")
dx = 2 * a
dy = np.sqrt(3) * a
i = count()
for y in range(ny):
offset = a if y % 2 == start else 0
for x in range(nx):
points[next(i)] = dx * x + offset, dy * y
return points
points = hex_pattern()
Now, we want some randomness in there. We set up several functions to add the randomness:
def linear(x: np.array, x0: float = -0.5, x1: float = 0) -> np.array:
"""
Linear gradient between x0 and x1, values lower x0 are set to 0,
higher than x1 to 1
"""
x = x.copy()
zeros = x < x0
ones = x > x1
z = x[(x >= x0) & (x <= x1)]
r = z.max() - z.min()
z = (z - z.min()) / r
x[(x >= x0) & (x <= x1)] = z
x[zeros] = 0
x[ones] = 1
return x
def norm_interval(x: np.array, a: float = -1, b: float = 1) -> np.array:
"""Normalize values to the new interval [a, b]"""
r = x.max() - x.min()
return (((x - x.min()) / r) * (b - a)) + a
def add_random(p: np.array, fun, k: float = 1, coord: int = 0) -> np.array:
"""Add a random displacement to a point with a threshold function"""
r = k * np.random.random(p.shape)
# The idea here is to scale the x (or y) values of the points to [-1, 1],
# then give it to the threshold function, which returns scaling factors [0, 1]
c = fun(norm_interval(p[:, coord])).reshape(-1, 1)
return p + r * c
points = add_random(points, linear)
Now, we can use the Voronoi function to generate the cells (or you use the points directly in the FreeCAD script):
vor = Voronoi(points)
plt.figure(figsize=(10,5))
voronoi_plot_2d(vor, plt.gca(), show_points=False, show_vertices=False)
plt.axis('equal')
plt.xlim(np.min(points[:, 0]), np.max(points[:, 0]))
plt.ylim(np.min(points[:, 1]), np.max(points[:, 1]))
plt.show()
And here we are:
vmdb2 is the new version of vmdebootstrap. It can create disk images, for example for Raspberry PIs or VMs. You could also use it as a debian installer, if you like...
However, there was is a shortcomming in vmdb2: it can only create raw images - but what you eventually want for VMs is qcow2. However, when you install this patch (actually, only the changes around line 92 are required), you can do the following to install onto a qcow2 image directly:
qemu-img create -f qcow2 myimage.qcow2 40G
modprobe nbd max_part=8
qemu-nbd --connect=/dev/nbd0 --format=qcow2 --cache=none --detect-zeroes=on --aio=native myimage.qcow2
vmdb2 --image /dev/nbd0 config.vmdb2
qemu-nbd --disconnect /dev/nbd0
Note however, that you must not use the kpartx module in the vmdb2 script.
For this purpose, it is not necessary because nbd will detect the partitions
automatically.
A minimal vmdb script would for example be:
steps:
- mklabel: msdos
device: "{{ image }}"
- mkpart: primary
device: "{{ image }}"
start: 10M
end: 100%
tag: rootfs
- mkfs: ext4
partition: rootfs
options: -E lazy_itable_init=0,lazy_journal_init=0
- mount: rootfs
- debootstrap: bullseye
mirror: http://deb.debian.org/debian
target: rootfs
- apt: install
packages:
- linux-image-amd64
tag: rootfs
- grub: bios
tag: rootfs
image-dev: "{{ image }}"